Pathway Disruption - to perform systematic disruption of specific protein-protein interactions in signaling networks that are related in diseases.
In mammalian cells, a given input (ligand) usually triggers many different signaling responses in different cell types. The decision as to which response to trigger in a certain cell type is made by a subtle and precise signaling mechanism yet to be understood. Traditionally, strategies to treat diseases caused by abnormal signaling have relied on blocking of ligand binding to corresponding receptors. However, this approach has a drawback of significant side effects due to a system-wide shutdown of the receptor-dependent pathways. For efficient treatment of diseases, therefore, it is desired to develop a strategy to surgically block the disease-related branch(es) of signaling cascades by identifying and disrupting specific protein-protein interactions.
Recent efforts on proteomics analyses have accumulated a vast data set of protein networks inside a cell and begun to elucidate precise information about linkages of protein-protein interactions in various disease states. This information allows to pinpoint the specific protein interactions downstream of receptors that are critical for the onset and progress of diseases of interest. For example, an unusual activation of JNK1 MAPK in JIP-1-dependent manner has been implicated in the oncogenic transformation precursor B-cells caused by the Bcr-Abl oncogene.12 Because of their important role in disease processes including cancer, inflammation and immune disorders, MAPK signaling components are important targets for therapeutic intervention. Especially, scaffold-mediated interactions can be a novel therapeutic target for surgical disruption of a specific subset of MAPK signaling.
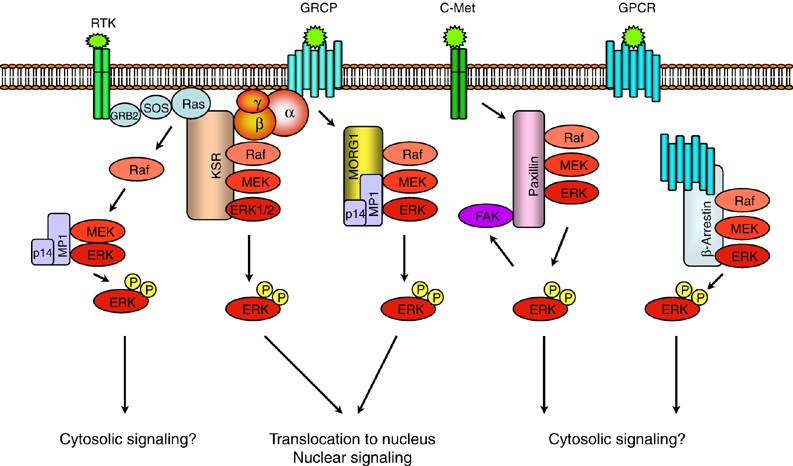
I will primarily focus on the mammalian MAPK scaffolds, JIP-1 and KSR-1 from the JNK and the ERK MAPK pathways, respectively. I propose to systematically disrupt the scaffold-kinase interactions in such diseased cells. In my previous study, I developed a novel genetic selection to screen an encoded combinatorial peptide library for dissociative inhibitors of a protein-protein interaction of interest.13 This selection strategy is amenable to ELISA-base assays for a high-throughput screen. I propose to apply this selection strategy to the systematic disruption of the JIP-1 and KSR-1 scaffold interactions with their component kinases. The positive isolates from selection will be tested for their ability to disrupt the target interactions in vitro and to block the target signaling pathways in vivo. The blocking of signaling by the dissociative inhibitors will be evaluated by monitoring its impact on the progress of disease states of interest. This strategy of pathway disruption by specific inhibition of critical protein interactions will be a valuable tool for evaluation of certain interactions for their role in disease development as well as the rapid generation of therapeutic agents for intervention of the disease itself.
