Regarding this topic, our group is currently focussing on two main areas:
I. A positive feedforward loop triggered by SHORTROOT shapens phloem sieve element development in the root
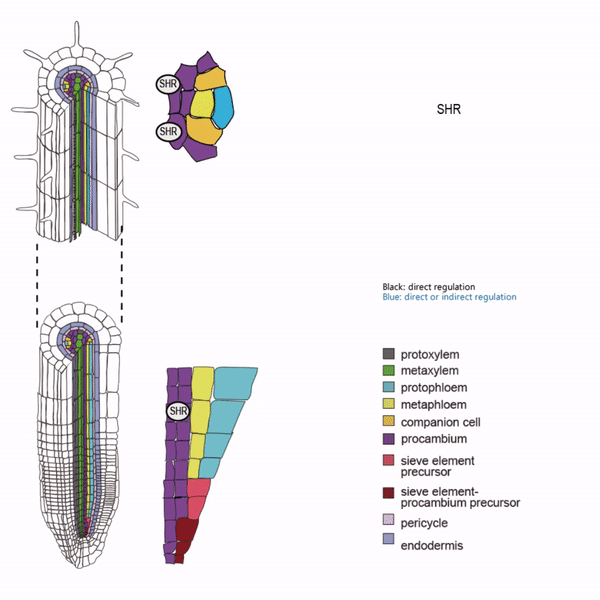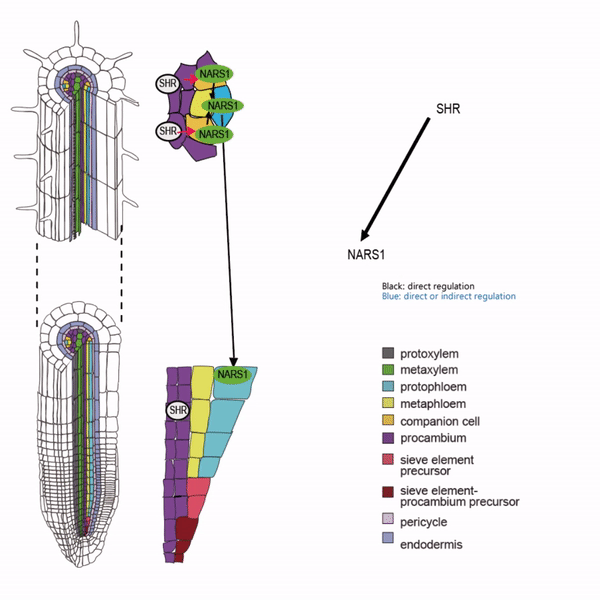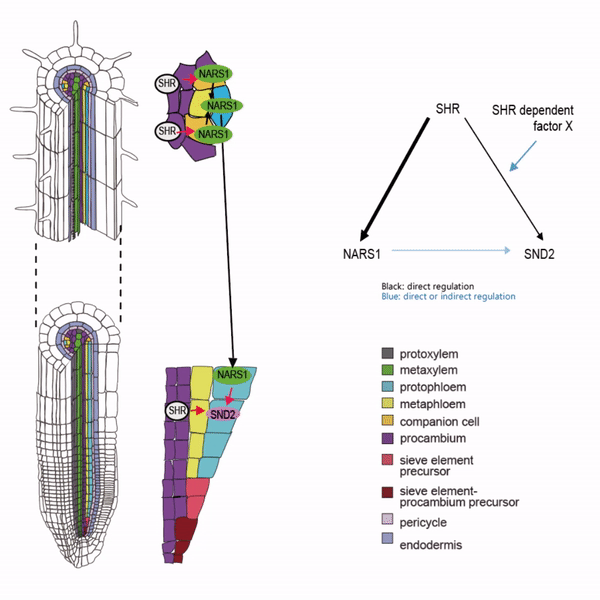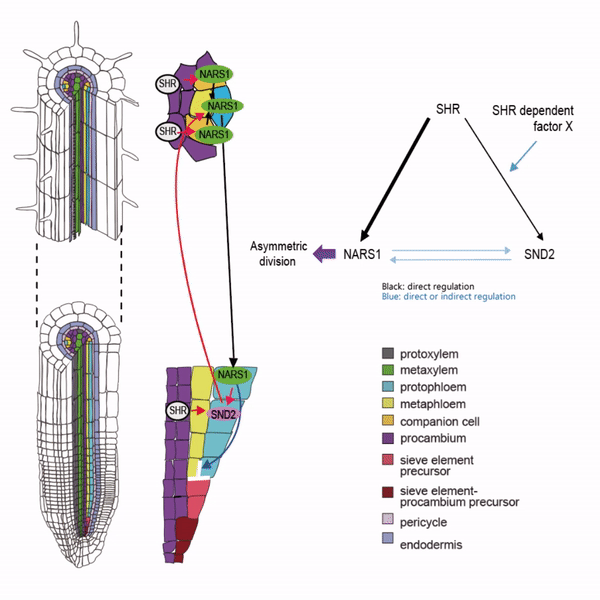
Sieve elements and companion cells constitute critical parts of the phloem tissue. These cells in the Arabidopsis root meristem arise via a series of asymmetric cell divisions of provascular cells. And, emergence of sieve elements and companion cells is highly coordinated. We recently published a paper in The Plant Cell, reporting how the phloem tissue is established in the root meristem (Kim and Zhou et al., 2020). In this, SHORTROOT (SHR), a master regulator of root development, is required for the asymmetric cell divisions for the phloem development. Companion cell formation is coordinated via the microRNA 165/6 activated by SHR that moves into the endodermis, while sieve element formation is via SHR moving into the phloem pole from the procambium. SHR in the phloem pole directly activates NARS1 and SND2, two NAC domain transcription factors. This regulation results in the amplification of a signal for asymmetric cell division, which seems to require for the movement of NARS1 from the differentiated part of the phloem towards the meristem.
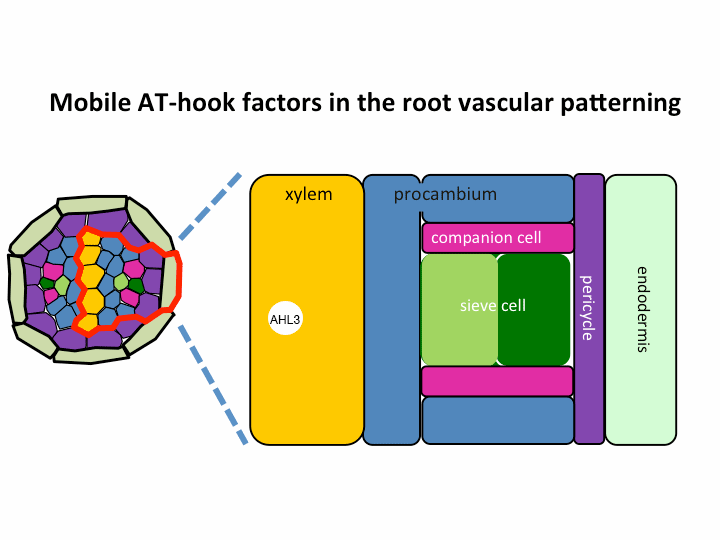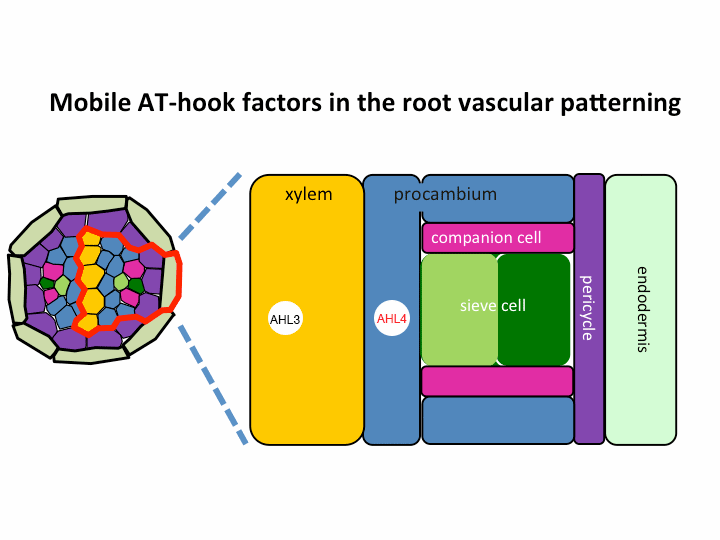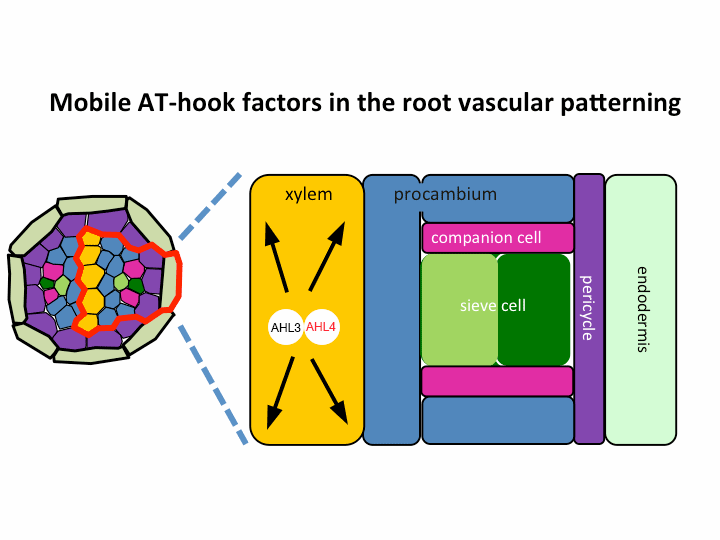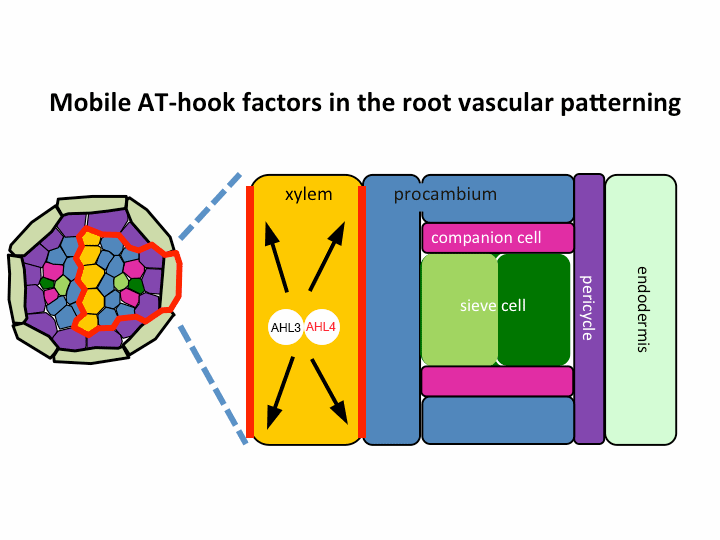
II. Investigation of AT-hook transcription factors that set up the xylem domain in Arabidopsis root
We previously reported that the intercellular movement of transcription factors is required for the proper vascular tissue patterning (Zhou et al., 2013). AHL3 and 4, two AT-hook family transcription factors play important roles in regulating the boundaries between xylem and procambium. AHL4 proteins are produced in the procambial cells in the root apical meristem and then move to xylem precursors. Without their movement, the expansion of xylem into procambium was observed. AHL4 plays this role by forming a protein complex with AHL3 (Figure below). AHL4 protein interacts with AHL3. To further understand the meaning of protein interactions in this process, we have been investigating whether AHL3 and 4 interact with other AT-hook like transcription factors in the gene family and whether these interactors are involved in the regulation of xylem development.