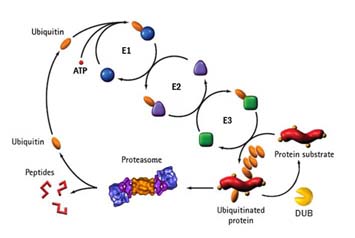
Ubiquitin and ubiquitination/ubiquitinylation
Ubiquitin is a ubiquitous protein, as the name implies, that is expressed in all eukaryotes ever known. It is small protein which is composed of 76 amino acids and conjugated to other proteins covalently which process is called as “ubiquitination or ubiquitinylation.”
There are so great many of ubiquitination substrates ; almost all physiology of cell has to do with protein ubiquitination and many disease are caused by ill-performance of ubiquitination. Even the conjugated ubiquitin itself is a substrate of ubiquitination, that is, covalently conjugated chain of ubiquitin is formed in the cell. So there is poly- ubiquitinated proteins as well as mono-ubiquitinated proteins and free ubiquitin.
Function of protein ubiquitination
As most post-translational protein modification, ubiquitination makes a change on protein function or physiology. Most well-known of function of protein ubiquitination is that of poly-ubiquitination which is linked via 48th lysine residue of ubiquitin. The K48 poly-ubiquitinated protein is recognized by the the protein degradation complex, 26S proteasome and specifically degraded and the ubiquitin monomer is recycled.
Mechanism of protein ubiquitination
Ubiquitination is mediated by E1-E2-E3 enzymatic reaction cascade, which is Ubiquitin Activation - Ubiquitin conjugation – Ubiquitin ligation. There is only one ubiquitin activation enzyme and there are more than 34 ubiquitin conjugating enzymes and 500 ubiquitin ligases in human. The Nobel Prize 2005 was awarded to A. Hershko, I. Rose, A. Ciechanover for their contribution on elucidation of E1-E2-E3 mechanism
Deubiquitination
Like de-phosphorylation, covalently conjgated ubiquitin is de-conjugated from the protein substrate. the enzyme called “Deubiquitinating enzymes(Dubs)” catalyses this chemical reaction. DUBs also processes Ub precursors (UCHs) and Ub adducts(UBPs). There are four families of enzymes on de-conjugation. (UCHs, UBPs, Otubain Family, JAMM family)
-------------------------------------------------------------------------------------------
Ubiquitin (Ub) is covalently attached to target proteins by a cascade enzyme system consisting of Ub activating (E1), conjugating (E2), and ligating (E3) enzymes. Ub E3 ligases that confer the substrate specificity have been grouped into two families: the HECT-domain family that is defined by its homology to E6-associated protein (E6AP) and the RING family carrying RING-finger domain that is essential for the Ub ligase activity. One of the well-defined RING E3 ligases is the Skp1/Cul1/F-box protein (SCF) complex, in which Cul1 serves as a scaffold molecule that interacts with Skp1 and a small RING-finger protein Roc1, also known as Hrt1 and Rbx1. F-box proteins are recruited to the complex by binding to the Skp1 adaptor protein.
At least six Cul members have been identified: Cul1, Cul2, Cul3, Cul4A, Cul4B, and Cul5. Of these, Cul3 is known to mediate the degradation of several proteins, such as cyclin E, but the molecular composition of Cul3-based Ub ligase was unknown. Recently, a large family of proteins having BTB (Bric-a-brac/ Tramtrack/Broad-complex) domain has been identified as novel Cul3-interacting proteins. Most BTB proteins, but not all, have additional domains for protein-protein interaction, such as Zn fingers, Kelch repeats, and MATH motifs. Furthermore, a subset of proteins containing BTB domain has been identified to function as substrate-specific adaptors that bind to Cul3. Specifically, MEL-26, a homolog of human SPOP (speckle-type POZ protein) in C. elegans, was first identified as BTB protein that serves as a specific adaptor of MEI-1 for the ubiquitination by Cul3-based Ub ligase and subsequent degradation by the proteasome. MEI-1 is a subunit of the katanin-like microtubule severing heterodimer MEI-1/MEI-2 that localizes to the spindles and the chromosomes during meiosis. SPOP BTB protein has also been shown to mediate the ubiquitination of the Polycomb group BMI and the variant histone MacroH2A . In addition, Keap1 BTB protein was shown to recruit Nrf2 to Cul3. Nrf2 is a transcription factor that regulates the expression of anti-oxidant genes upon oxidative stress. In S. pombe, Btb1p, Btb2p, and Btb3p interact with Cul3 but their functions remain unknown. Therefore, so far only a few protein substrates have been shown to interact with BTB proteins for their ubiquitination by Cul3-based Ub ligases. Daxx was originally identified as a protein that binds to the death domain of Fas receptor by yeast two-hybrid screening. Daxx interacts with the apoptosis signal-regulating kinase 1 (ASK1) and promotes Fas-mediated apoptosis through the activation of Jun N-terminal kinase (JNK). Subsequent studies have shown that Daxx behaves as a pro-apoptotic protein under various stress conditions. On the contrary, homozygous deletion of the Daxx gene has been shown to cause extensive apoptosis and embryonic lethality, suggesting that Daxx is an anti-apoptotic protein. Recent studies also support that Daxx is anti-apoptotic rather than pro-apoptotic.
Daxx also plays a role as a transcription regulator by interacting with various nuclear proteins involved in transcription. Daxx interacts with HDAC-II, core histones, and a chromatin-associated protein, Dek, suggesting that Daxx represses basal transcription through chromatin remodeling. Daxx has also been shown to form a ternary complex with homeodomain proteins Pax-3 and Pax-7 to inhibit transcription, while Pax-3/FKHR fusion protein in alveolar rhabdomyosarcoma cells is resistant to Daxx inhibition. Similarly, Daxx interacts with ETS1 to repress transcriptional activation of the MMP1 gene. It has also been demonstrated that Daxx suppresses cell death induced by p53 over-expression and p53-dependent stress response, thus acting as a negative regulator of p53. Interestingly, the level of Daxx protein has been shown to decrease in HeLa cells upon induction of apoptosis by actinomycin D or exposure to UV. Moreover, Daxx has recently been shown to bind to SPOP. These reports suggest a possibility that the functions of Daxx can be regulated by proteolysis.
We demonstrate that SPOP recruits Daxx to Cul3 and promotes the ubiquitination of Daxx and subsequent degradation by the proteasome. Moreover, SPOP/Cul3-mediated degradation of Daxx reversed Daxx-mediated repression of ETS1- and p53-dependent transcription. We also demonstrate that Daxx degradation triggers apoptosis. Taken together, we suggest that SPOP/Cul3-Ub E3 ligase plays an essential role in the control of Daxx level and thus in the regulation of Daxx-mediated cellular processes, including transcriptional regulation and apoptosis.
Resently, other data suggest a possibility that ubiquitination/degradation of Daxx can be reversed by USP. Further research should elucidate the stabilization of Daxx by USP and other mechanisms that regulate Daxx transcriptional repression activity are under work including sumoylation and phosphoryaltion of Daxx.
