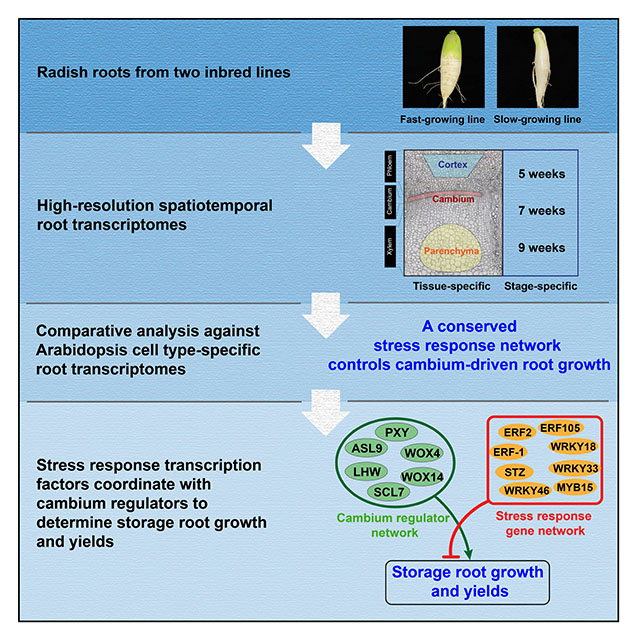
Cambium is the key meristem that drives the growth of stems and roots in lateral direction (also referred to as thickening) in vascular plants and determines biomass and yields in root crops. To find novel regulatory programs driving the cambium-driven secondary growth, we recently employed radish as exemplary root crop and generated high-resolution spatiotemporal transcriptome data using Laser Capture Microdissection (Hoang, Choe and Zhen et al., 2020). Transcriptome dynamics in different tissues and growth stages of radish inbred lines with contrasting storage root growth and yields provided us with extensive information about potential players in the secondary growth. To further narrow down key networks, we performed conserved co-expression network analyses by comparing radish data with combined Arabidopsis root data published by Zhang et al. (2019), and thereby identified well-conserved networks that include several transcription factors previously characterized to be involved in stress responses. The involvement of these stress response transcription factors in cambium-driven secondary growth is not well-understood. By studying the in vivo networks composed of these transcription factors and well-known canonical cambium regulators, we found that these two subnetworks coordinately interact with each other to determine cambium-driven growth and yields. The interaction between these two gene regulatory subnetworks is further supported by published data for major root crops (Hoang et al., 2020).