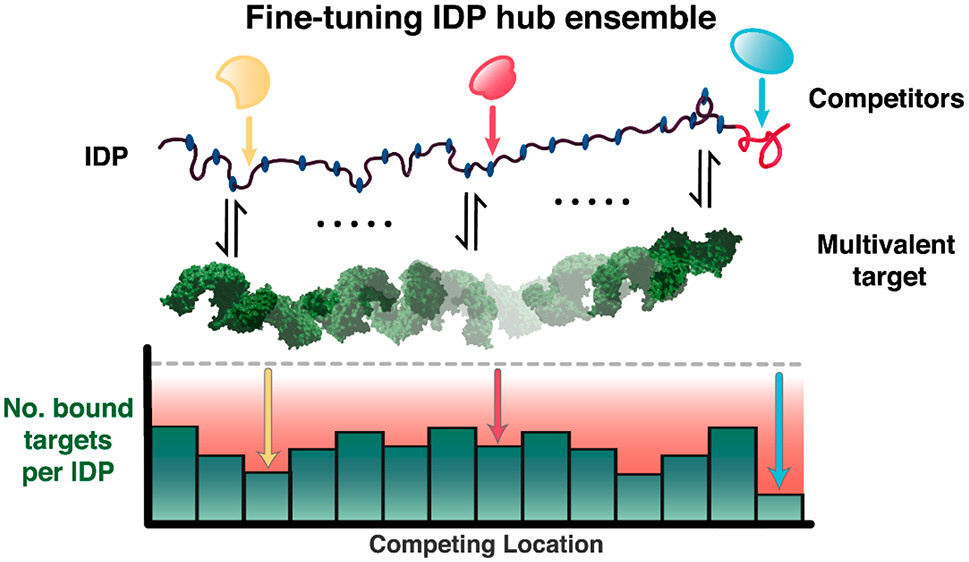
A central theme of our research is to elucidate the molecular mechanisms governing fundamental biological processes by quantitative characterization of the underlying macromolecular interactions. In particular, we use the concepts and methods from biophysical chemistry (structure, thermodynamics, and kinetics) to scrutinize the multi-component interactions involved in two different systems: Nuclear Pore Complex (NPC) in nucleocytoplasmic transport and Mediator Complex in transcriptional regulation. A remarkable feature of these molecular machines is that many of their protein subunits, collectively termed Nups in the NPC and Meds in the Mediator Complex, contain intrinsically disordered regions (IDRs) dynamically interconverting among multiple conformational states. Our specific aim is to explore novel regulatory roles of these IDRs in the following processes:
1) Assembly of the NPC and the Mediator Complex from their ~30 distinct subunits.
2) Interactions of the NPC with nucleocytoplasmic transport receptors and of the Mediator complex with transcription factors.
3) Conformational changes in these complexes coupled to the interactions with their binding partners.
In parallel, we aim to devise cellular systems to probe the effects of such conformational changes on nucleocytoplasmic transport and transcription in vivo.
Given the abundance of IDRs in the eukaryotic proteome, the molecular principles and quantitative methods established in the proposed research will be applicable to other complex biological systems. These basic studies will also provide invaluable information for future medical investigations, culminating in innovative therapeutic approaches targeting IDRs in human diseases.
생물리화학 실험실
Laboratory of Biophysical Chemistry
생물리화학 연구실